noncoding-RNA-seq¶
What it does¶
The snakePipes noncoding-RNA-seq workflow allows users to process their single or paired-end ribosomal-depleted RNA-seq fastq files upto the point of gene/transcript/repeat-element counts and differential expression. Repeat elements are quantified and tested for differential expression at the name, family and class level. Since changes in repeat element expression tend to be unidirectional, size factors from gene expression are used when normalizing repeat element expression.
Note that in addition to the normal GTF file, this pipeline requires a repeat masker output file, which can be downloaded from UCSC or other sites. The chromosome names here must match that used in the other files.
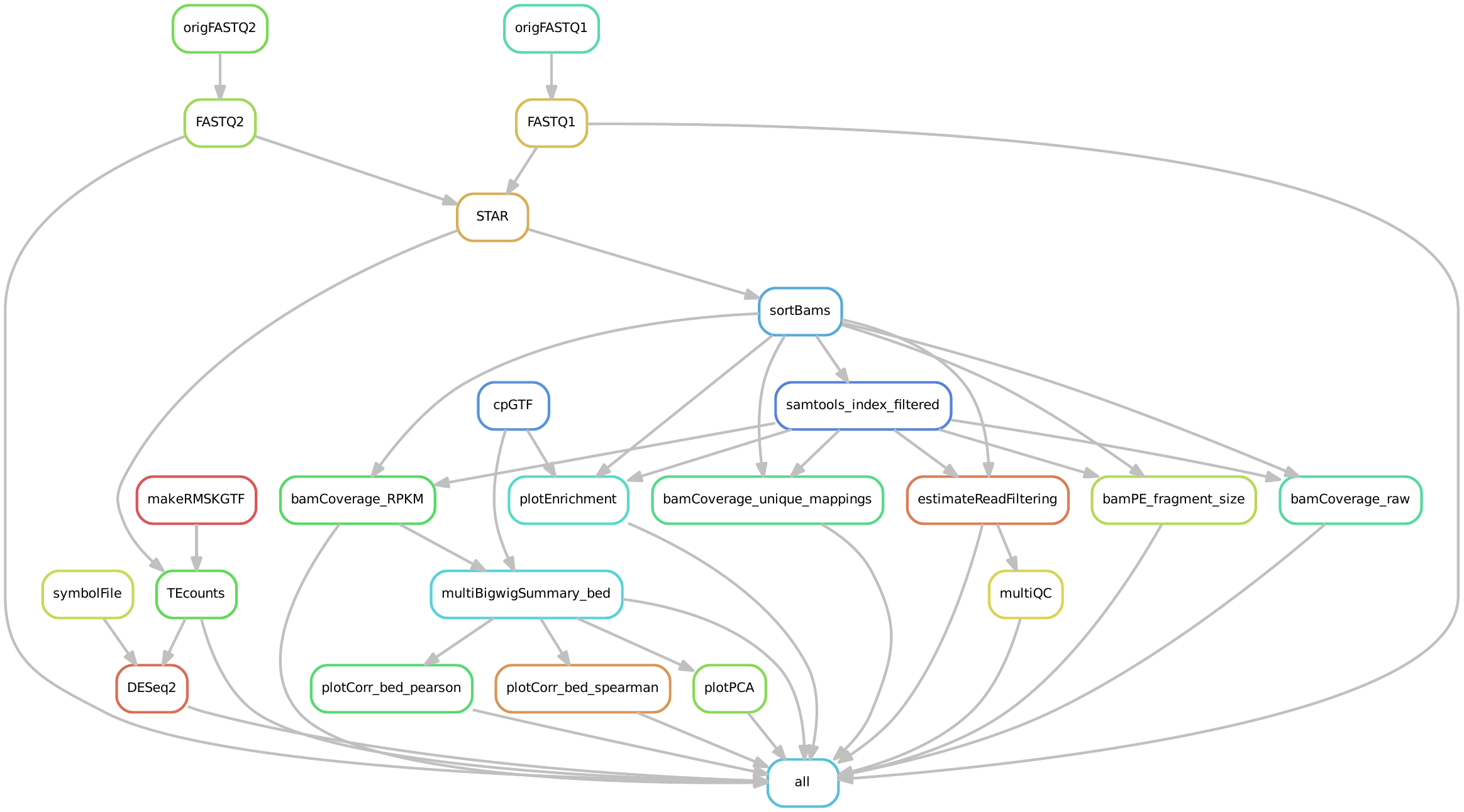
Input requirements¶
The only requirement is a directory of gzipped fastq files. Files could be single or paired end, and the read extensions could be modified using the keys in the defaults.yaml file below.
Configuration file¶
There is a configuration file in snakePipes/workflows/noncoding-RNA-seq/defaults.yaml:
## General/Snakemake parameters, only used/set by wrapper or in Snakemake cmdl, but not in Snakefile
pipeline: noncoding-rna-seq
outdir:
configFile:
clusterConfigFile:
local: False
maxJobs: 5
## directory with fastq files
indir:
## preconfigured target genomes (mm9,mm10,dm3,...) , see /path/to/snakemake_workflows/shared/organisms/
## Value can be also path to your own genome config file!
genome:
## FASTQ file extension (default: ".fastq.gz")
ext: '.fastq.gz'
## paired-end read name extension (default: ["_R1", "_R2"])
reads: ["_R1","_R2"]
## assume paired end reads
pairedEnd: True
## Number of reads to downsample from each FASTQ file
downsample:
## Options for trimming
trim: False
trimmer: fastp
trimmerOptions:
## further options
mode: alignment,deepTools_qc
sampleSheet:
bwBinSize: 25
fastqc: False
fragmentLength: 200
libraryType: 2
## supported mappers: STAR HISAT2
aligner: STAR
alignerOptions: "--outSAMstrandField intronMotif --outFilterMultimapNmax 1000 --outFilterMismatchNoverLmax 0.1 --outSAMattributes Standard"
verbose: False
plotFormat: png
#### Flag to control the pipeline entry point
fromBAM: False
bamExt: '.bam'
#umi_tools
UMIBarcode: False
bcPattern: NNNNCCCCCCCCC #default: 4 base umi barcode, 9 base cell barcode (eg. RELACS barcode)
UMIDedup: False
UMIDedupSep: "_"
UMIDedupOpts: --paired
Apart from the common workflow options (see Running snakePipes), the following parameters are useful to consider:
aligner: The only choice at the moment is STAR.
alignerOptions: Options to pass on to your chosen aligner. Note that library type and junction definitions don't have to be passed to the aligners as options, as they are handeled either automatically, or via other parameters.
plotFormat: You can switch the type of plot produced by all deeptools modules using this option. Possible choices : png, pdf, svg, eps, plotly
Differential expression¶
Like the other workflows, differential expression can be performed using the --sampleSheet option and supplying a sample sheet like that below:
name condition
sample1 eworo
sample2 eworo
SRR7013047 eworo
SRR7013048 OreR
SRR7013049 OreR
SRR7013050 OreR
Note
The first entry defines which group of samples are control. This way, the order of comparison and likewise the sign of values can be changed. The DE analysis might fail if your sample names begin with a number. So watch out for that!
Complex designs with blocking factors¶
If the user provides additional columns between 'name' and 'condition' in the sample sheet, the variables stored there will be used as blocking factors in the order they appear in the sample sheet. Eg. if the first line of your sample sheet looks like 'name batch condition', this will translate into a formula batch + condition. 'condition' has to be the final column and it will be used for any statistical inference.
Multiple pairwise comparisons¶
The user may specify multiple groups of independent comparisons by providing a 'group' column after the 'condition' column. This will cause the sample sheet to be split into the groups defined in this column, and a corresponding number of independent pairwise comparisons will be run, one for each split sheet, and placed in separate output folders named accordingly. This will be applied to DESeq2 pairwise comparison. Specifying a value of 'All' in the 'group' column will cause that sample group to be used in all pairwise comparisons, e.g. if the same set of controls should be used for several different treatment groups.
An example sample sheet with the group information provided looks like this:
name condition group sample1 Control All sample2 Control All sample3 Treatment Group1 sample4 Treatment Group1 sample5 Treatment Group2 sample6 Treatment Group2
Analysis modes¶
Following analysis (modes) are possible using the noncoding-RNA-seq workflow:
"alignment"¶
In this mode, the pipeline uses STAR to create BAM files and TEtranscripts to quantify genes and repeat elements. Differential expression of genes and repeat elements is then performed with DESeq2.
"deepTools_qc"¶
The pipeline provides multiple quality controls through deepTools, which can be triggered using the deepTools_qc mode. It's a very useful add-on with any of the other modes.
Note
Since most deeptools functions require an aligned (BAM) file, the deepTools_qc mode will additionally perform the alignment of the fastq files. However this would not interfere with operations of the other modes.
Understanding the outputs¶
Assuming the pipline was run with --mode 'alignment,deepTools_qc' on a set of FASTQ files, the structure of the output directory would look like this (files are shown only for one sample)
├── bamCoverage
│ ├── sample1.coverage.bw
│ ├── sample1.RPKM.bw
│ ├── sample1.uniqueMappings.fwd.bw
│ ├── sample1.uniqueMappings.rev.bw
├── cluster_logs
├── deepTools_qc
│ ├── bamPEFragmentSize
│ │ ├── fragmentSize.metric.tsv
│ │ └── fragmentSizes.png
│ ├── estimateReadFiltering
│ │ └── sample1_filtering_estimation.txt
│ ├── logs
│ │ ├── bamPEFragmentSize.err
│ │ ├── bamPEFragmentSize.out
│ │ ├── multiBigwigSummary.err
│ │ └── plotCorrelation_pearson.err
│ ├── multiBigwigSummary
│ │ └── coverage.bed.npz
│ ├── plotCorrelation
│ │ ├── correlation.pearson.bed_coverage.heatmap.png
│ │ ├── correlation.pearson.bed_coverage.tsv
│ │ ├── correlation.spearman.bed_coverage.heatmap.png
│ │ └── correlation.spearman.bed_coverage.tsv
│ ├── plotEnrichment
│ │ ├── plotEnrichment.png
│ │ └── plotEnrichment.tsv
│ └── plotPCA
│ ├── PCA.bed_coverage.png
│ └── PCA.bed_coverage.tsv
├── DESeq2_sampleSheet
│ ├── DESeq2_report_genes.html
│ ├── DESeq2_report_repeat_class.html
│ ├── DESeq2_report_repeat_family.html
│ ├── DESeq2_report_repeat_name.html
│ ├── DESeq2.session_info.txt
│ ├── genes_counts_DESeq2.normalized.tsv
│ ├── genes_DEresults.tsv
│ ├── genes_DESeq.Rdata
│ ├── repeat_class_counts_DESeq2.normalized.tsv
│ ├── repeat_class_DEresults.tsv
│ ├── repeat_class_DESeq.Rdata
│ ├── repeat_family_counts_DESeq2.normalized.tsv
│ ├── repeat_family_DEresults.tsv
│ ├── repeat_family_DESeq.Rdata
│ ├── repeat_name_counts_DESeq2.normalized.tsv
│ ├── repeat_name_DEresults.tsv
│ └── repeat_name_DESeq.Rdata
├── FASTQ
├── filtered_bam
│ ├── sample1.filtered.bam
│ ├── sample1.filtered.bam.bai
├── multiQC
├── STAR
└── TEcount
└── sample1.cntTable
Note
The _sampleSheet suffix for the DESeq2_sampleSheet is drawn from the name of the sample sheet you use. So if you instead named the sample sheet mySampleSheet.txt then the folder would be named DESeq2_mySampleSheet. This facilitates using multiple sample sheets.
Apart from the common module outputs (see Running snakePipes), the workflow would produce the following folders:
bamCoverage: This would contain the bigWigs produced by deepTools bamCoverage . Files with suffix
.coverage.bware raw coverage files, while the files with suffixRPKM.bware RPKM-normalized coverage files.deepTools_QC: (produced in the mode deepTools_QC) This contains the quality checks specific for mRNA-seq, performed via deepTools. The output folders are names after various deepTools functions and the outputs are explained under deepTools documentation. In short, they show the insert size distribution(bamPEFragmentSize), mapping statistics (estimateReadFiltering), sample-to-sample correlations and PCA (multiBigwigSummary, plotCorrelation, plotPCA), and read enrichment on various genic features (plotEnrichment)
DESeq2_[sampleSheet]: (produced only if a sample-sheet is provided.) The folder contains the HTML result reports DESeq2_report_genes.html, DESeq2_report_repeat_name.html, DESeq2_report_repeat_class.html and DESeq2_report_repeat_family.html as we as the annotated output file from DESeq2 (genes_DEresults.tsv, etc.) and normalized counts for all samples, produced via DEseq2 (genes_counts_DESeq2.normalized.tsv, etc.) as well as an Rdata file (genes_DESeq.Rdata, etc.) with the R objects
dds <- DESeq2::DESeq(dds)andddr <- DDESeq2::results(dds,alpha = fdr). Sample name to plotting shape mapping on the PCA plot is limited to 36 samples and skipped otherwise.filtered_bam: This contains sorted and indexed BAM files that have been filtered to remove secondary alignments. This are used by deepTools and are appropriate for use in IGV.
multiQC: This folder contains the report produced by MultiQC and summarizes alignment metrics from STAR and possibly various deepTools outputs.
STAR: This would contain the output logs of RNA-alignment by STAR. The actual BAM files are removed at the end of the pipeline as they're not compatible with typical visualization programs
TEcount: (produced in the alignment mode) This contains the counts files and logs from the TEcount program in the TEtranscripts package. These are used by DESeq2 for differential expression.
Command line options¶
MPI-IE workflow for noncoding RNA mapping and analysis
- usage example:
noncoding-RNA-seq -i input-dir -o output-dir mm10
usage: noncoding-RNA-seq -i INDIR -o OUTDIR [-h] [-v] [--ext EXT]
[--reads READS READS] [-c CONFIGFILE]
[--clusterConfigFile CLUSTERCONFIGFILE] [-j INT]
[--local] [--keepTemp]
[--snakemakeOptions SNAKEMAKEOPTIONS] [--DAG]
[--version] [--emailAddress EMAILADDRESS]
[--smtpServer SMTPSERVER] [--smtpPort SMTPPORT]
[--onlySSL] [--emailSender EMAILSENDER]
[--smtpUsername SMTPUSERNAME]
[--smtpPassword SMTPPASSWORD] [-m MODE]
[--downsample INT] [--trim]
[--trimmer {cutadapt,trimgalore,fastp}]
[--trimmerOptions TRIMMEROPTIONS] [--fastqc]
[--bcExtract] [--bcPattern BCPATTERN] [--UMIDedup]
[--UMIDedupSep UMIDEDUPSEP]
[--UMIDedupOpts UMIDEDUPOPTS] [--bwBinSize BWBINSIZE]
[--plotFormat STR] [--aligner ALIGNER]
[--alignerOptions ALIGNEROPTIONS]
[--sampleSheet SAMPLESHEET] [--fromBAM]
[--bamExt BAMEXT] [--singleEnd]
GENOME
Positional Arguments¶
- GENOME
Genome acronym of the target organism. Either a yaml file or one of: hg38, mm39_ens106, GRCh38_gencode40, dm6, GRCz10, SchizoSPombe_ASM294v2, mm10_gencodeM19, GRCz11
Required Arguments¶
- -i, --input-dir
input directory containing the FASTQ files, either paired-end OR single-end data
- -o, --output-dir
output directory
General Arguments¶
- -v, --verbose
verbose output (default: 'False')
- --ext
Suffix used by input fastq files (default: '".fastq.gz"').
- --reads
Suffix used to denote reads 1 and 2 for paired-end data. This should typically be either '_1' '_2' or '_R1' '_R2' (default: '['_R1', '_R2']). Note that you should NOT separate the values by a comma (use a space) or enclose them in brackets.
- -c, --configFile
configuration file: config.yaml (default: 'None')
- --clusterConfigFile
configuration file for cluster usage. In absence, the default options specified in defaults.yaml and workflows/[workflow]/cluster.yaml would be selected (default: 'None')
- -j, --jobs
maximum number of concurrently submitted Slurm jobs / cores if workflow is run locally (default: '5')
- --local
run workflow locally; default: jobs are submitted to Slurm queue (default: 'False')
- --keepTemp
Prevent snakemake from removing files marked as being temporary (typically intermediate files that are rarely needed by end users). This is mostly useful for debugging problems.
- --snakemakeOptions
Snakemake options to be passed directly to snakemake, e.g. use --snakemakeOptions='--dryrun --rerun-incomplete --unlock --forceall'. WARNING! ONLY EXPERT USERS SHOULD CHANGE THIS! THE DEFAULT VALUE WILL BE APPENDED RATHER THAN OVERWRITTEN! (default: '['--use-conda']')
- --DAG
If specified, a file ending in _pipeline.pdf is produced in the output directory that shows the rules used and their relationship to each other.
- --version
show program's version number and exit
Email Arguments¶
- --emailAddress
If specified, send an email upon completion to the given email address
- --smtpServer
If specified, the email server to use.
- --smtpPort
The port on the SMTP server to connect to. A value of 0 specifies the default port.
- --onlySSL
The SMTP server requires an SSL connection from the beginning.
- --emailSender
The address of the email sender. If not specified, it will be the address indicated by --emailAddress
- --smtpUsername
If your SMTP server requires authentication, this is the username to use.
- --smtpPassword
If your SMTP server requires authentication, this is the password to use.
Options¶
- -m, --mode
workflow running modes (available: 'alignment, deepTools_qc') (default: '"alignment,deepTools_qc"')
- --downsample
Downsample the given number of reads randomly from of each FASTQ file (default: 'False')
- --trim
Activate fastq read trimming. If activated, Illumina adaptors are trimmed by default. Additional parameters can be specified under --trimmerOptions. (default: 'False')
- --trimmer
Possible choices: cutadapt, trimgalore, fastp
Trimming program to use: Cutadapt, TrimGalore, or fastp. Note that if you change this you may need to change --trimmerOptions to match! (default: '"fastp"')
- --trimmerOptions
Additional option string for trimming program of choice. (default: 'None')
- --fastqc
Run FastQC read quality control (default: 'False')
- --bcExtract
To extract umi barcode from fastq file via UMI-tools and add it to the read name (default: 'False')
- --bcPattern
The pattern to be considered for the barcode. 'N' = UMI position (required) 'C' = barcode position (optional) (default: '"NNNNCCCCCCCCC"')
- --UMIDedup
Deduplicate bam file based on UMIs via umi_tools dedup that are present in the read name. (default: 'False')
- --UMIDedupSep
umi separation character that will be passed to umi_tools.(default: '"_"')
- --UMIDedupOpts
Additional options that will be passed to umi_tools.(default: '')
- --bwBinSize
Bin size of output files in bigWig format (default: '25')
- --plotFormat
Possible choices: png, pdf, None
Format of the output plots from deepTools. Select 'none' for no plots (default: '"png"')
- --aligner
Program used for mapping: STAR (default: '"STAR"'). If you change this, please change --alignerOptions to match.
- --alignerOptions
STAR option string, e.g.: '--twopassMode Basic' (default: '"--sjdbOverhang 100 --outSAMstrandField intronMotif --outFilterMultimapNmax 1000 --outFilterMismatchNoverLmax 0.1 --outSAMattributes Standard --outSAMunmapped Within --outSAMtype BAM Unsorted"')
- --sampleSheet
Information on samples (required for DE analysis); see 'https://github.com/maxplanck-ie/snakepipes/tree/master/docs/content/sampleSheet.example.tsv' for example. The column names in the tsv files are 'name' and 'condition'. The first entry defines which group of samples are control. This way, the order of comparison and likewise the sign of values can be changed. The DE analysis might fail if your sample names begin with a number. So watch out for that! (default: 'None')
- --fromBAM
Input folder with bam files. If provided, the analysis will start from this point. If bam files contain single ends, please specify --singleEnd additionally.
- --bamExt
Extention of provided bam files, will be substracted from basenames to obtain sample names. (default: '".bam"')
- --singleEnd
input data is single-end, not paired-end. This is only used if --fromBAM is specified.